
We recently advertised 16 new PhD positions within our newly founded Max Planck Research School for the Science of Human History, an institution tied both to the University of Jena and the Max Planck Institute for the Science of Human History in Jena.
Within this call, I am seeking candidates for working on a new project in my group titled “Co-estimating Human Mobility and Language Dispersal with Ancient DNA and Linguistic Data”
It is based on a scientific article that I learned about recently, by Lisa Loog and coworkers from University College London. In this paper, Loog and colleagues develop a new estimator for human mobility throug time, based on ancient genetic data. I want to briefly explain in this post how their estimation of human mobility works, and how I intend to extend it in this PhD project.
The challenge of analysing ancient genetic data is that data is distributed sparsely both in space and in time. For example, we might have samples from 4000 years ago from one region, and from 3000 years ago from another region, so what does their genetic relationship plausibly say about the movement of people through time and space?
Loog and colleagues addressed this challenge in a new way. They realised the following simple principle: If people move around a lot, different ancestries tend to come together close in space. In contrast, if people tend to stay where they are born for many generations, neighbours tend to have similar ancestries. Put in their own words:
If mobility was low (i.e., strong spatial structure), then we would expect differences between entities to be more strongly correlated with space, relative to time, whereas if mobility was high we would expect time to explain a relatively larger proportion of differences between entities (because of the homogenizing effects of high mobility across space).
So it is clear that in general, the closer people are in space and time, the more genetically similar they should be. But the relative weights of how similar they are in space versus time is a direct estimator for human mobility.
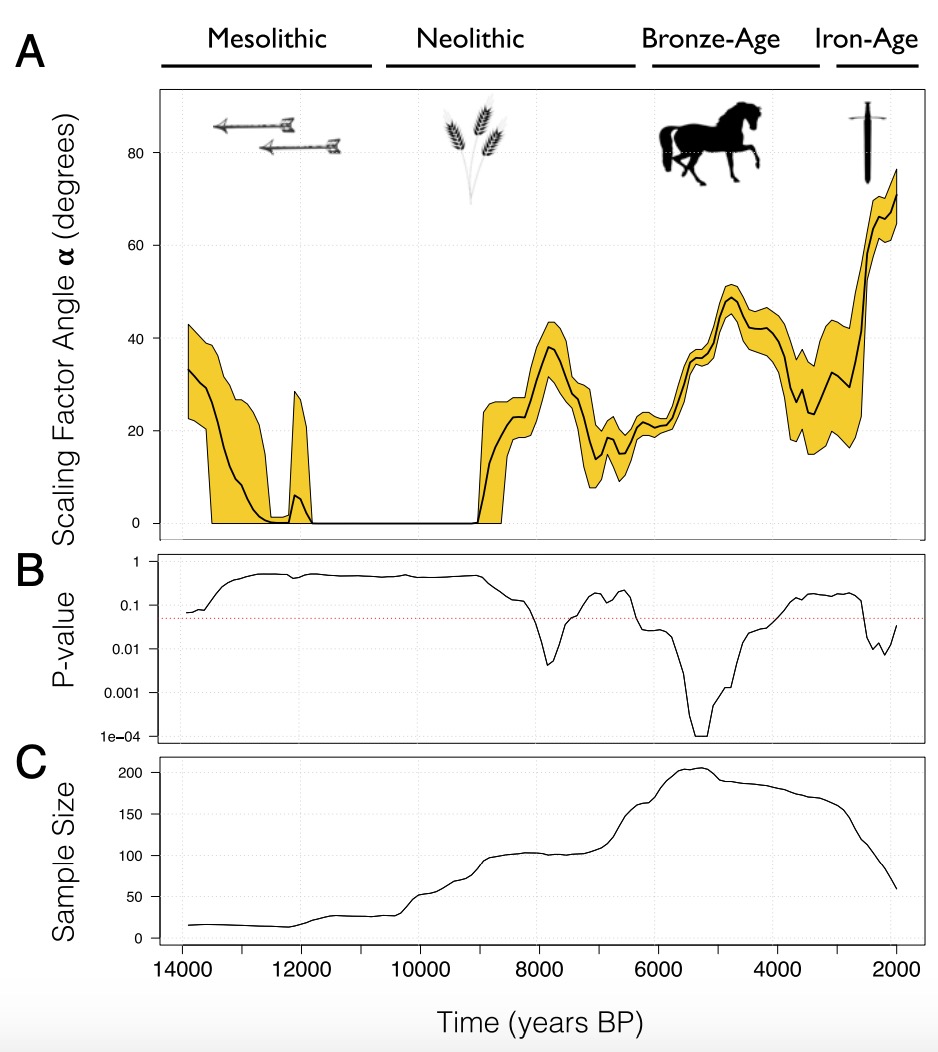
Loog and colleagues used this basic idea to develop a statistical method that tracks human mobility through time. Here is their main figure:

This analysis covers West Eurasia, and it shows an estimate of human mobility from 14,000 years ago until 2,000 years ago. The yellow band in panel A describes the uncertainty of their migration rate (mobility) estimator through time. The first decay between 14,000 and 12,000 years ago can be ignored for now, as it is not statistically significant. However there are three spikes of human mobility in more recent times that correspond strikingly to several key transformation periods in human history.
The first time where human mobility peaked is between 8,000 and 6,000 years ago, which is the time when agriculture first spread through Europe, as a new way of life, and — as genetic studies have shown — as new people who descend from early Anatolian farmers and who first replaced and then intermixed with indigenous Hunter-Gatherers all throughout Europe. The second period of peaking human mobility was between around 5,000 and 3,500 years ago, according to this analysis, which corresponds to the onset of the Bronze Age and is the time of another large migration in Europe, this time with ancestry from the Pontic Caspian Steppe. The third peak in their analysis falls into the Iron Age and perhaps later.
This striking description of human migration through time was quantified using human data directly, rather than from archaeological or historical sources, which in contrast to genetic data are somewhat more open to interpretation.
OK, so what else is there to do in this PhD project? One thing that we are good at in our IMPRS is the combination of different disciplines, such as Linguistics and Genetics. In particular, I would like to use the Loog et al. framework to investigate language dispersal on top of human mobility.
Of course, languages spread differently from genes, and while the spread of languages and genes is often correlated, it doesn’t have to. Takes as an example England: While England has undergone a 100% Language replacement in the early Medieval times (from Celtic to Germanic), there certainly was not a 100% replacement of people (our estimate of admixture is 40%, and that is actually already much higher than what many other scholars believe).
So how can we combine linguistic and genetic data in a combined model inspired by Loog et al.? My idea is to introduce linguistic distances between samples, much as we have genetic distances. The simplest one could just be a probability for having spoken different languages. Such a metric is difficult to assign, but it could be done with some uncertainty.
With a dataset consisting of pairwise genetic distances as well as linguistic distances, we can then try to jointly estimate human mobility and language dispersal. Of course, there are lots of caveats to be figured out, but with high risk comes high gain: We might learn quantitatively, how much people carried around their languages as they moved, or to what extent they adopted existing local languages.
If you are looking to pursue a PhD, not afraid of mathematical modelling, some programming, and working with (very different kinds of) data, then consider applying to this project!