
Human history is deeply imprinted in our genomes, through patterns of common ancestry. These patterns can be studied by looking at mutations that are shared between people living in different times in different places. In this post I will summarize our recent genetic study on Anglo-Saxon immigration history in Britain, and in doing so,will describe some new methodology that we have developed for this project, which is based on rare mutations.
A large majority of the inhabitants of Great Britain and Ireland today speak English, a language related to Dutch and German. Going back 2,000 years, their native tongue was a Celtic language, related to Welsh, Gaelic and Irish, which are spoken by relatively few people today compared to English.
This change of language is a striking example for the large cultural impact of the Anglo-Saxon migration period, which started around 400AD with peoples from the continental coast (mainly Jutes, Angles, Saxons and Frisians) migrating into the South and East of England. From archaeology, we know that during this period burial practices, jewellery industry, religious rituals and many aspects of communal life, changed.
Who were the Anglo-Saxons? How did they interact with the local population? Today, we can find answers to these questions from Genetics, in particular with the help of ancient DNA.
We collected teeth from 8 female and 2 male individuals who lived in villages near Cambridge at various times between 2,000 and 1,200 years ago.. From the teeth we drilled bone powder, extracted hundreds of millions of short fragments of DNA and by sequencing these fragments we reconstructed the first complete ancient genomes from Great Britain to date (another study with ancient British genomes came out at the same time).
From precise carbon-dating, we know that the ten ancient genomes can be divided into two groups: Three individuals are from the late Iron Age, around 100BC-50AD, and 7 samples are from the Early and Middle Anglo-Saxon period (450–750AD). We were eager to see whether there were systematic genetic differences between these two age groups and between each of them and modern English individuals.
The first thing we noticed is that all ten individuals are genetically relatively similar, and that they all looked relatively “English”, similar to modern English individuals. This was expected, because the indigenous (Celtic) population in Britain and the immigrating (Germanic) population were very closely related, with a common ancestor perhaps a few hundred generations back in time. Most methods to analyse population relationships are based common mutations, most of which are shared between all populations in the world today, so it is hard to use them to distinguish two recently diverged populations in small Europe.
We therefore decided to look for rare mutations that are shared by only a few people out of hundreds of people. The general idea is that if we see a mutation that is shared among two isolated populations, this mutation must have occurred at a time when those two populations were still together, so before their isolation. The longer two populations have been isolated, the fewer shared mutations between these two populations we should see. This effect is most pronounced for rare mutations, because rare mutations are relatively younger than common mutations, so are more informative about recent population separations.
To find these rare mutations, we analysed the genomes of hundreds of people living today in Europe and selected mutations that were found in not more than 0.5% of all chromosomes. Because we were interested in finding “Anglo-Saxon-specific” mutations, we especially looked for genetic variants found in modern-day Dutch people (who live closest to where the Anglo-Saxons supposedly came from). We then looked at how many of those rare “Dutch mutations” were shared by our ancient samples and by modern British people. Here is a figure showing the results:
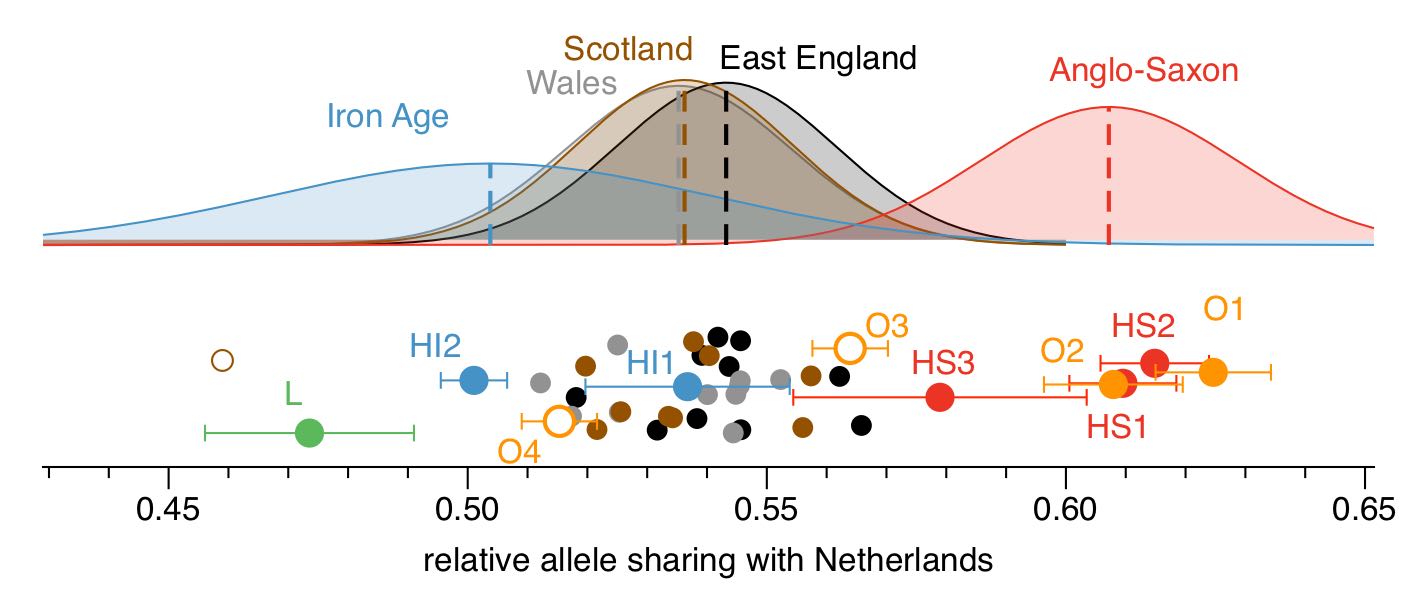
Note that the absolute numbers shown below the plot are normalized and without absolute meaning. Only the relative positioning of the samples matters. First, we can see that most of the Anglo-Saxon samples (in red and orange) carry about 50% more rare Dutch mutations than the Iron Age samples do (in blue and green), which is quite a strong signal. Second, we find that modern English samples carry an intermediate number of rare Dutch mutations, compared to Iron Age and Anglo-Saxon samples. More precisely, the modern English samples place at 38% on average between the Iron Age and the Anglo-Saxon samples (100% would be all the way to the right at the Anglo-Saxons, 0% would be all the way left to the Iron Age samples). And because allele sharing is linear under admixture, we conclude that modern English can be modelled as a two-way mixture of 38% Anglo-Saxon ancestry, and 62% Iron Age ancestry.
When I say “most of the Anglo-Saxon samples”, I meant that we had two exceptions. Two samples, labeled O3 and O4 in the figure above stand out: Sample O3 places somewhere in between the Anglo-Saxon and Iron Age samples, while sample O4 falls completely within the range of the Iron Age samples. This suggested to us that O3 and O4 carry 50% and 100% indigenous ancestry, respectively.
At this point it is worth pointing out that O1, O2, O3 and O4 are four female samples excavated in Oakington (East England), on a large Anglo-Saxon cemetery with remains from over 120 individuals. Archaeologically, that cemetery is clearly of Anglo-Saxon culture. So the two suspicous women (O3 and O4) were buried with Anglo-Saxon grave goods and alongside other Anglo-Saxons (O1 and O2). It was therefore very surprising for us to find that two out of four individuals that we looked at where of (partial) indigenous British ancestry. It suggests that at least in this one cemetery, mixing and inter-marriage between immigrants and Natives occured already at the very first phase of the Anglo-Saxon period.
There is one important caveat to our way of modelling modern England as a two-way mixture: We assume that the Iron-Age samples represent “Pre-Anglo-Saxon” England, although there were about 400 years between the Iron Age and until the Anglo-Saxons came. And this was the time period where the Roman empire ruled the country. However, any external genetic contribution from Roman time would only influence our mixture estimate if it was equally close to the Dutch as the Anglo-Saxons were. We think that any such contribution was — if at all — small. A similar argument holds for other later continental contributions from the Vikings or Normans. Fortunately, another ancient DNA study confirms this view by genetically analysing ancient British soldiers from the Roman time, and found that there was overall genetic continuity during the Roman occupation.
I think the Anglo-Saxon migration period is a prime example for a relatively recent event in human history that is now made accessible to genetic studies. In this post I have summarized some of the main results of our work on that subject using ancient DNA. In the next post I will cover our new method rarecoal, which allows us to infer population history more explicitly from rare mutations, and also to pin down the relationship of the ancient samples with modern Europeans more quantitatively.
Further reading:
Coverage on BBC News: English DNA ‘one-third’ Anglo-Saxon
Our original article: Schiffels, S. et al. Iron Age and Anglo-Saxon genomes from East England reveal British migration history. Nat. Commun. 7:10408 doi: 10.1038/ncomms10408 (2016).
The other article on ancient British DNA: Martiniano, R. et al. Genomic signals of migration and continuity in Britain before the Anglo-Saxons. Nat. Commun. 7:10326 doi: 10.1038/ncomms10326 (2016).